- Home
-
Screening
- Ionic Screening Service
-
Ionic Screening Panel
- Ligand Gated Ion Channels
- Glycine Receptors
- 5-HT Receptors3
- Nicotinic Acetylcholine Receptors
- Ionotropic Glutamate-gated Receptors
- GABAa Receptors
- Cystic Fibrosis Transmembrane Conductance Regulators (CFTR)
- ATP gated P2X Channels
- Voltage-Gated Ion Channels
- Calcium Channels
- Chloride Channels
- Potassium Channels
- Sodium Channels
- ASICs
- TRP Channels
- Other Ion Channels
- Stable Cell Lines
- Cardiology
- Neurology
- Ophthalmology
-
Platform
-
Experiment Systems
- Xenopus Oocyte Screening Model
- Acute Isolated Cardiomyocytes
- Acute Dissociated Neurons
- Primary Cultured Neurons
- Cultured Neuronal Cell Lines
- iPSC-derived Cardiomyocytes/Neurons
- Acute/Cultured Organotypic Brain Slices
- Oxygen Glucose Deprivation Model
- 3D Cell Culture
- iPSC-derived Neurons
- Isolation and culture of neural stem/progenitor cells
- Animal Models
- Techinques
- Resource
- Equipment
-
Experiment Systems
- Order
- Careers
Childhood Absence Epilepsy
Children absence epilepsy (CAE) is a systemic idiopathic epilepsy, characterized by frequent typical absence seizures, bilateral symmetric synchronized 3 Hz rhythmic spike slow wave discharge. In children, the prevalence rate is 0.4/1 000 ~ 0.7 /1 000, accounting for about 5% ~ 15% of childhood epilepsy; the incidence rate is 5.8 / 100 000 ~ 7. 1/100 000, girls The incidence rate is higher than that of boys; the onset is usually between 4 and 10 years old, and the peak age is 5 to 7 years old.
Genetic Characteristics
The influence of genetic factors in the process of CAE is indispensable, but its inheritance is still not completely clear.
Since 1961, some scholars have begun to study the EEG of children with CAE, and found that genetic factors play an important role in CAE. The disease is autosomal dominant with age-dependent manifestations. Later research by Berkovic et al. found that CAE is not simply a single-gene inheritance, but a complex disease affected by multiple independent or independent genetic factors, which belongs to the category of multi-gene inheritance. In recent years, genetic research on CAE has emerged in an endless stream, and the mechanisms discovered have become more and more abundant, including susceptible gene mutations and multi-gene genetic mutations. This suggests that the genetic mechanism of CAE is very complex and needs to be comprehensively viewed from many aspects.
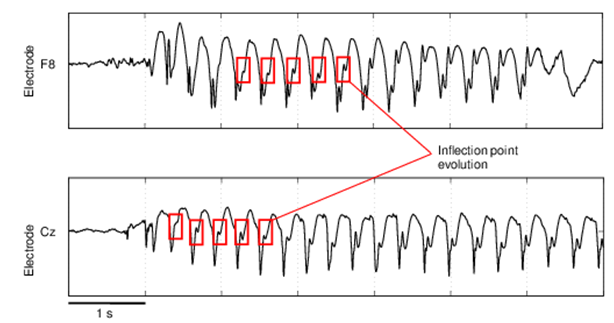
Figure 1.Scalp EEG from human subjects with childhood absence epilepsy. (Serafim Rodrigues, et al.; 2010 )
Pathogenesis
The genetic mechanism of CAE is complex. In addition to the mutations of some susceptible genes that have been identified by the academic community, the currently discovered mechanisms are genome fragment copy number variations (CNVs).
Susceptibility gene mutation
There are many susceptibility genes in CAE, and a considerable part of them are related to coding ion channels. Several susceptibility genes closely related to CAE have been found so far. Mutations of the same susceptibility gene can also be found in other related epilepsy syndromes, such as febrille seizure (FS), generalized epilepsy with febrile seizures (GEFS+), etc.
Calcium Channel
Calcium ion channel is a hotspot in the traditional research of CAE susceptibility genes. Voltage-dependent calcium channels can be divided into T-type, P/Q-type, N-type, R-type and L-type according to their pharmacology and voltage sensitivity. The mechanism of causing epilepsy may be related to the rhythm disorder of the thalamus cortex Related, and the onset of CAE is related to the excitability of the thalamus cortical system.
GABAA Receptor Related Genes
GABAA receptor-related genes are another major class of susceptibility genes that are closely related to CAE. GABAA receptors are composed of α1, α2, β1, β2, γ, and δ subunits. GABAA receptors have an inhibitory effect on neurons, so the number, structure or function of GABAA receptors is impaired, which can increase the susceptibility to epilepsy. The mechanism may be related to the generation of spike wave discharge. Recently, studies have found that the single cell genetic expression of the GABAA receptor α1 subunit mutation in a mouse model will change the formation of dendritic spines and GABAergic nodules, inhibit the signal transmission of GABAergic synapses, and thus trigger CAE.
GABRA6 is another GABAA receptor gene related to CAE. Hernandez and other studies have shown that mutations in GABRA6 will reduce the current density of the GABAA receptor, shorten the channel opening time and the duration of the pulse, thus affecting the synthesis of the GABAA receptor and increasing the susceptibility of CAE.
In addition to the above genes, Feucht and others passed the multi-allelic transmission imbalance test and found that the GABRB3 gene is also highly correlated with CAE. Studies have found that mutations in GABRB3 will disrupt the normal upper membrane of the GABAA receptor β3 subunit and hinder its maturation , Influencing the inhibitory function of the GABAA receptor, which triggers CAE. In addition, the GABRG2 gene encodes the GABAA receptor γ2 subunit, and its mutations will down-regulate the expression level of the γ2 subunit on the cell surface, making the γ2 subunit and other subunits Problems with the assembly, which damages the structure of GABAA receptors on neurons.
NOTCH1 Gene
The latest research shows that the expression of the NOTCH1 gene and its homologue NLE1 gene, which have an important role in brain development, can activate the related mRNA and reduce the production of GFAP, thereby reducing the number of glial cells. The increased ratio of neuronal cells to glial cells will affect the nutrition and repair function of neurons after damage, which is related to the incidence of CAE. In addition, researcheres also found that the application of NOTCH1 antagonists, that is, γ-secretase inhibitor (DAPT) can promote the generation of epilepsy-like discharges, proving the relationship between the NOTCH signaling pathway and CAE from the negative side.
CNVs
In addition to the above-mentioned mutation mechanisms of related susceptibility genes, in recent years, CNVs (including deletion, insertion or replication of gene fragments) have also been found to be related to the pathogenesis of CAE. As an important genetic structure variation, CNVs directly or indirectly affect changes in gene expression through changes in its copy number and transcriptional regulatory factors, thereby causing phenotypic differences among individuals. Studies have shown that 9.3% of patients with GGEs have CNVs. A genomic fragment can contain multiple genes, so the impact of CNVs on CAEs is more complicated than the pathogenic mechanism of susceptible genes. Some CAE-related CNVs overlap with CAE's susceptibility gene loci, such as the NIPA2 gene at 15q11.2 locus. However, many CNVs beyond the susceptibility gene locus have also been found.
References
- Crunelli V, Leresche N. Childhood absence epilepsy: genes, channels, neurons and networks. Nat Rev Neurosci. 2002, 3(5):371-382.
- Avoli, M., et al.; Generalized epileptic disorders: an update. Epilepsia. 2002, 42, 445–457.
- Commission on Classification and Terminology of the International League Against Epilepsy. Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia 30, 389–399 (1989).
- Serafim Rodrigues, et al.; A method for detecting false bifurcations in dynamical systems: Application to neural-field models. Biological Cybernetics. 2010, 102(2):145-54.
Related Section
Inquiry

